Parasites and Microbes
Overview
We apply our decades of genomic experience and extensive collaborative networks of universities, research groups and health organisations to study how people’s immune systems and microbiomes (the communities of microbes in their guts and lungs) respond to infections and the environmental conditions of where they live (climate change, transport links and medical intervention).
Together with our scientific partners, we are establishing a global network of sentinel sites to investigate infectious disease in context. This includes how pathogens evolve and spread, the underlying genetics of silent (symptomless) carriage, long-range transmission, and how the microbiome affects infectious disease susceptibility, at the local, national and international level.
How we have contributed to infectious disease research
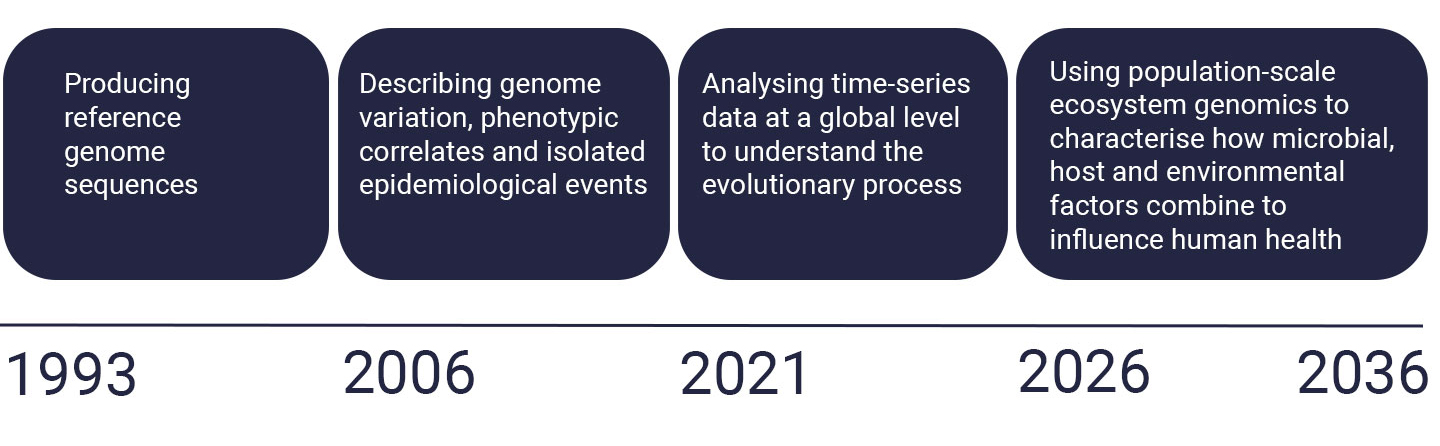
Global research hub
Since its inception, the Parasites and Microbes programme has become a key global hub for pathogen genome research. Initially we delivered reference genomes for the bacteria responsible for a wide range of diseases and the applied this knowledge to understand the global spread and evolution of pandemic and epidemic infections.
Microbiome culturing and analysis
More recently our scientists have developed and apply metagenomic techniques and methods to culture and analyse microbial communities to explore the microbiome of people’s guts and lungs.
Empowering the world-wide research community
This work has enabled us to:
- build long-term national and international partnerships
- develop open data tools and datasets for the global research community
- guide disease control interventions
- forge close links with local and international health organisations for mutual benefit.
Building on this foundation of technical expertise, genetic research experience and equitable global research collaborations, we now seek to deliver population-scale ecosystem genomics to truly understand the interplay between all the factors (genetic, environmental and human) that affect disease severity, spread and drug resistance.
Our Aims
Looking beyond the genome – exploring infectious diseases in context
The factors that determine the severity and transmission of an infection are much more complex than simply the genetic makeups of the bacteria, virus, parasites, and human hosts involved. A wide range of factors act both independently and in combination to affect human health. These include:
- environmental issues (climate change)
- human-made infrastructure (transport links)
- medical interventions (vaccination programmes and antibiotics)
- people’s underlying genetics (disease susceptibility)
- the bacteria and viral communities in people’s guts and respiratory systems (microbiomes).
Yet, until now, research and preventative strategies have focused on the seeking to understand and address each of these factors separately.
Exploring the interplay of infectious disease factors in context and at high resolution
Advances in genomic techniques, coupled with our extensive and well-developed international collaborative research networks, now mean that we can investigate all aspects that affect disease severity and transmission directly in the places where people live. This give us an unprecedented opportunity to collect high-resolution longitudinal datasets to characterise how the following factors act as key modifiers of human health:
- pathogen
- microbiome
- immune
- demographic
- environmental.
Gathering and standardising the data needed for the full picture
For these reasons our research studies seek to couple microbial genomic or transcriptomic sequence data generated from host and environmental samples with:
- microbiological data (for example antimicrobial susceptibility testing)
- host serology and metabolomic data
- patient health records
- human genomic and demographic data (for example mobility)
- abiotic/biotic measurements of the environment in which the populations live.
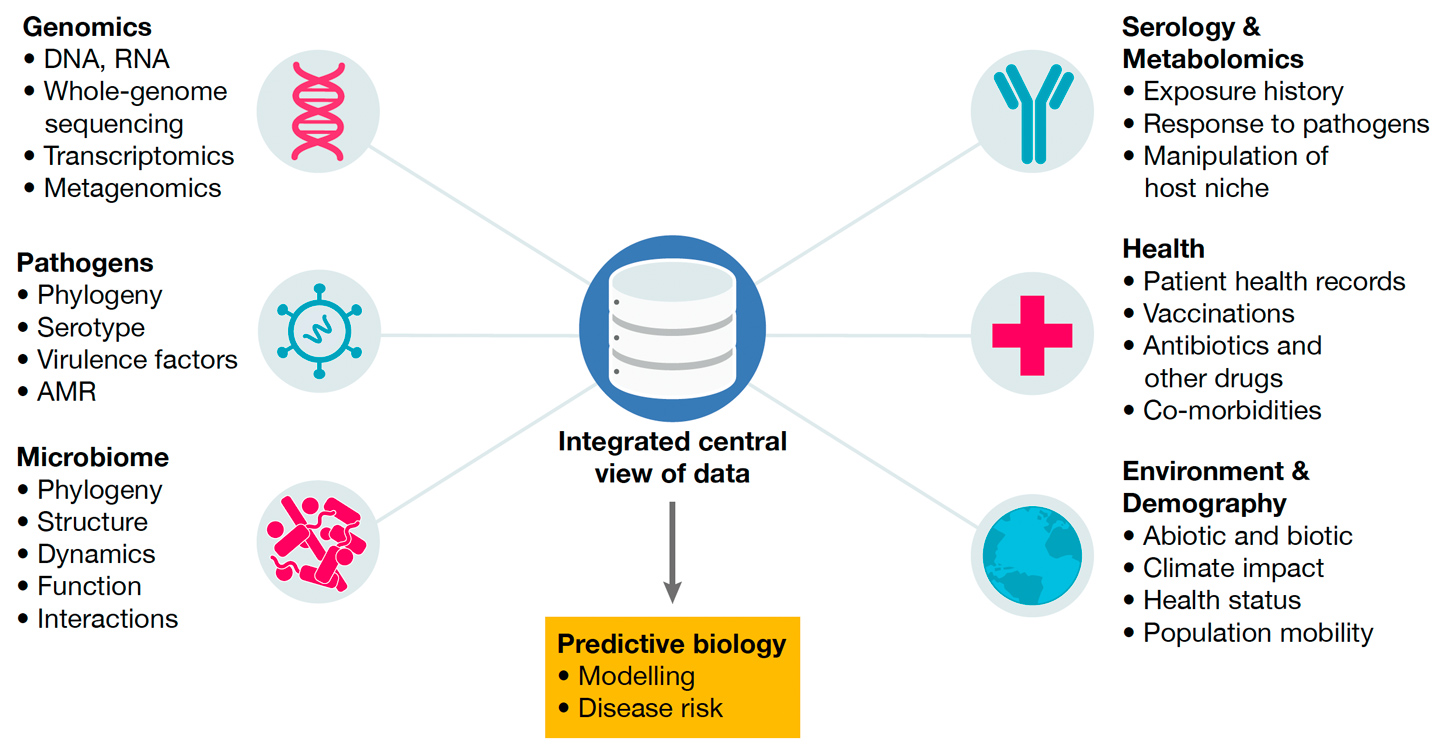
Linking genotypic insights to phenotypic consequences
Linking the genotypic insights gained through population-scale genomics with phenotypic consequences is vital to maximise the biological understanding generated from samples collected and genomic data generated. Accordingly, we are establishing high-throughput platforms for:
- phenotypic screening using imaging and machine learning to characterise abiotic and biotic perturbations on microbes (for example, antibiotics and pollutants)
- metabolomics to understand how microbes modify their environment
- systems serology (including serum immunoassays and B-cell sequencing) to determine how exposure to colonisation and infection affects the human immune system.
Positively impacting healthcare delivery and public health strategies around the world
Wherever possible we seek to centralise activities so that our researchers and our collaborators can capitalise on improvements in technology and data flow. Through this approach, we seek to empower our partners to establish platforms for routine clinical, community, and environmental sampling, that are to linked electronic health records and demographic data to maximise insight and impact.
Answering critical questions for preventing and treating disease
We investigate human populations as a whole, through sampling both healthy individuals and those presenting with disease, to address critical questions in understanding, preventing and treating disease. For example, we seek to discover:
- What are the human, environmental and microbial factors involved in determining whether an infection results in symptomless colonisation or disease?
- How does the co-circulation of multiple pathogens, or the interactions between pathogens, microbiomes and hosts contribute to seasonal outbreaks?
- How do these factors impact people’s ability to fight off infections (for example by diminishing our ability to fight off pathogens through phenomena such as antibody-dependent enhancement)?
The daily interplay of between these complex factors determine whether disease control interventions are likely to be effective or not, yet they are poorly understood. Our work seeks to change this.
Proactively identifying new and emerging pathogens
We apply metagenomic approaches to identify new and emerging viruses and bacteria from the samples our scientists and collaborative networks collect. By applying population-scale ecosystem genomics we will be able to compare the microbiome in different populations around the world to uncover emerging strains and hidden chains of transmission.
Creating truly representative knowledge to improve global healthcare – discovery research
Our research seeks to address gaps in data and knowledge to ensure greater access to the health benefits arising from a genomic view of health and disease.
In 2024, 71 per cent of all human microbiome data has been generated from populations living in Europe, the United States, and Canada. In contrast, less than 2 per cent of the data has come from populations in India, Pakistan, and Bangladesh. Yet these three countries represent one-quarter of the entire global population.
Addressing this disparity will provide a better understanding of what constitutes a ‘normal’ microbiome, improving our ability to positively impact population health worldwide.
Population-scale ecosystem genomics
Building on the foundation of our genomic technologies, bioinformatic techniques and equitable collaborative sampling networks we now seek to deliver population-scale ecosystem genomics. By integrating a wide range of datasets gathered at key global sites, our researchers and their collaborators can characterise how pathogen, microbiome, immune, demographic, and environmental factors combine to affect disease severity, spread, and drug resistance.
For example, we explore poorly understood, yet fundamental, phenomena such as asymptomatic colonisation (silent carriers), long-range transmission, and pathogen-microbiome-host interactions.
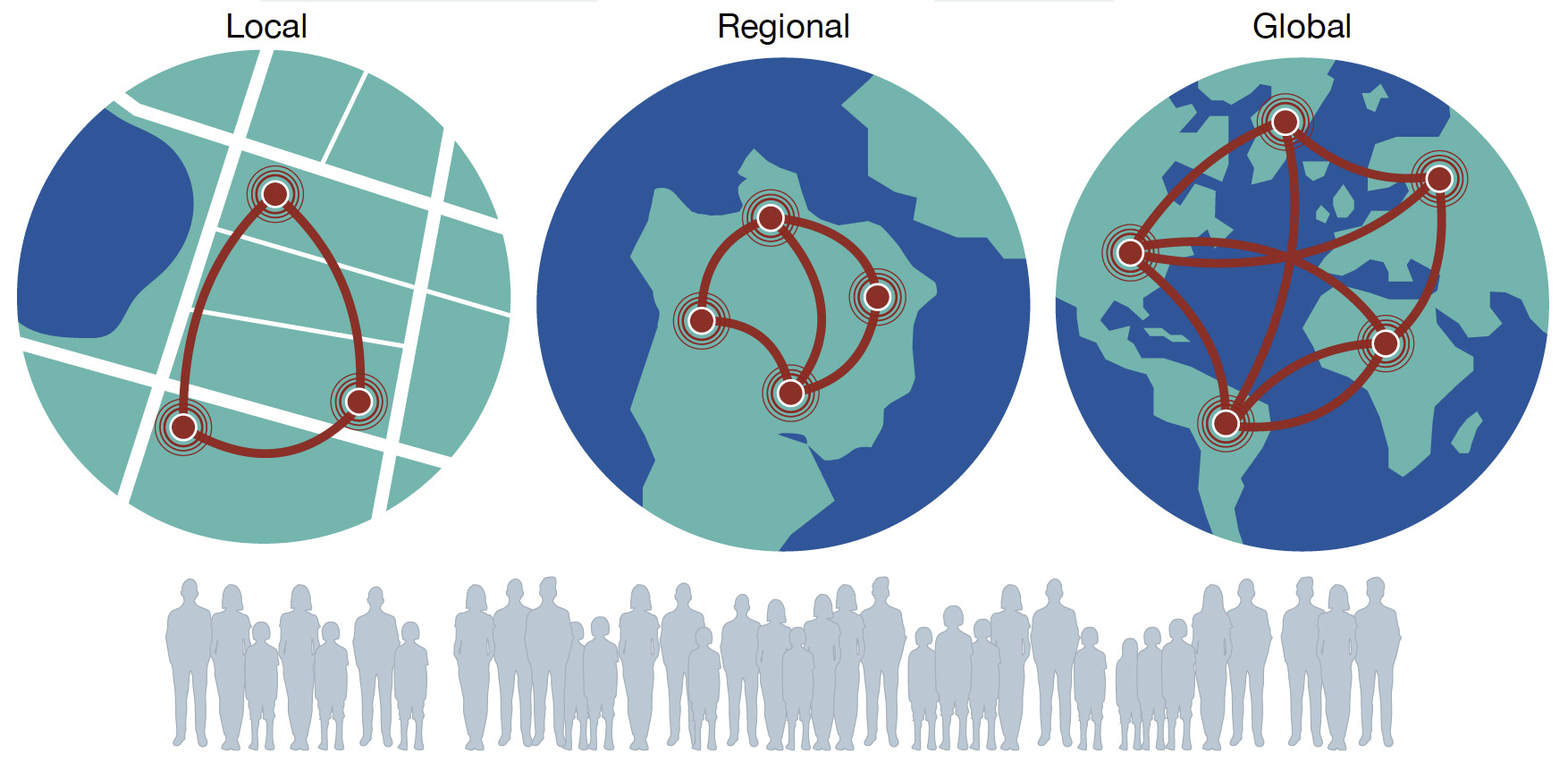
Population-scale sampling of healthy and infected individuals
Key drivers of how interactions between microbes, hosts and the environment can shape human health and disease often only become apparent when moving beyond the study of individuals or small cohorts. Accordingly, longitudinal sampling at population-scales, from local, national, regional to global, are crucial. Prospective sampling of healthy individuals, as well as those presenting with disease among these populations enables investigation of the poorly understood processes that impact many aspects of our daily lives and determine whether disease control interventions are likely to be effective.
Sentinel sites
We are combining the Sanger Institute’s scale and expertise in partnership with our extensive collaborative networks to co-develop population-scale longitudinal studies at ‘sentinel sites’. These sites cover health settings in high-, middle-, and low-income countries.
To create datasets of unparalleled richness and compatibility from within and between sentinel sites, we develop and use standardised approaches for:
- sample collection
- data generation
- integration
- analysis.
Working with sentinel sites maximises our ability to build genomic research capacity. It also offers opportunities to:
- reveal new understandings of the differences in disease risk and outcome
- explore the impact of disease control strategies
- study the evolution of antimicrobial resistance
- detail the effects of climate change
- deliver culturally and geographically sensitive insights
- develop translational products.
Integrating disparate data types in integrated central data resources
Working at population scales, through globally important sentinel sites and with other partners in our collaborative network provides an unprecedented opportunity to collect high-resolution longitudinal datasets to characterise how pathogen, microbiome, immune, demographic, and environmental factors act as key modifiers of human health.
To ensure that the disparate data types being generated can be used by all collaborators to extract valuable new insights, we seek to generate ever-more integrated central data resources and develop shared, scalable protocols. Our aim is that these resources and processes can be uniformly applied from high- to low-income settings to:
- standardise data generation
- reduce noise
- enable descriptions of complex relationships and interactions across population levels (local, national, international).
These integrated central data resources will ensure that data, biological insights and models, are freely and openly available within a trusted research environment. This will enable our scientists and our collaborators to compare between populations across sentinel sites, validate patterns observed in their individual datasets, and reveal complex relationships and interactions across different levels of a system.
Developing AI-enabled predictive disease models
The resulting rich and inter-compatible datasets will enable us to develop predictive disease models that answer fundamental scientific questions and will:
- provide insights that allow more accurately targeted interventions
- shorten the timeline for outbreak detection
- identify new strategies for reshaping the microbiome
- suggest ways to combat antimicrobial resistance
- offer routes to reduce vaccine evasion.
Building equitable research partnerships around the world to deliver real-world benefits
All our research is built on established long-term national and international networks that are design to provide benefit to all scientists and communities involved. These networks and collaborations include more than 300 research groups in 69 countries around the world.
We develop open data tools and datasets relevant to basic science and disease control, as well as close links with local and international health organisations. Wherever possible we seek to centralise activities shared with our strategic partners to deliver transformative biological insights that impact human health and wellbeing.
In partnership with our collaborators, we capitalise on improvements in technology and data flow to establish platforms for routine clinical, community, and environmental sampling, with linked electronic health records and demographic data.
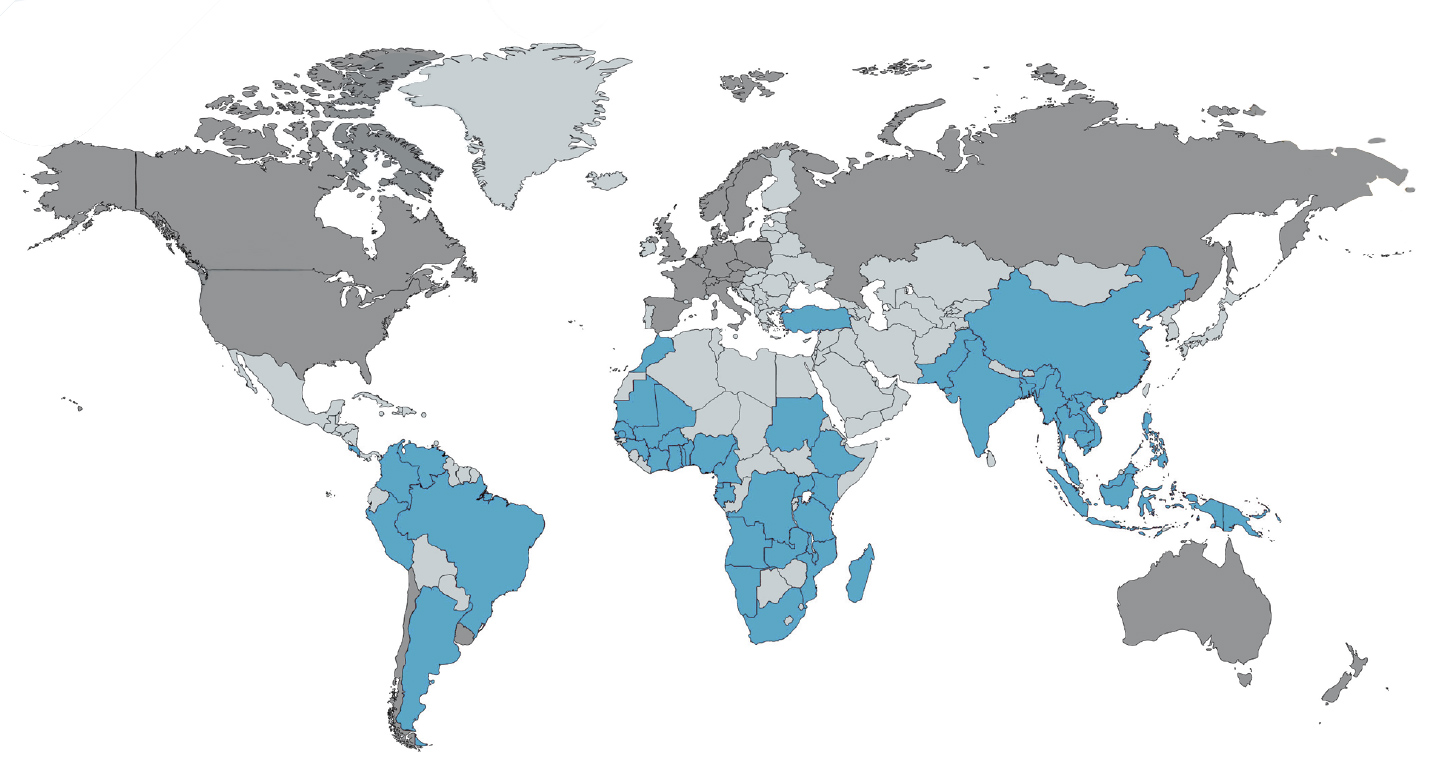
Extended research family
Our extensive collaborative research with scientists and public health bodies around the globe provides an ‘extended family’ that enables the programme to deliver impactful science at a scale well beyond the capacity of any individual research group within the programme.
Through our International Fellows, Associate Faculty, and Honorary Faculty, we have strong collaborative ties and strategic links with established and future global leaders in microbial genomics. In partnership with these key scientific thinkers, we seek to ensure that our research and its outputs deliver real-world impact both now and for many years to come.
Strategic relationships power translation into real-world benefits
In addition to our core and extended Faculty team, we have strong strategic partnerships and leverage the deep domain expertise that exists across the Sanger Institute, and a wide range of external organisations. In this way we can ensure that the insights and knowledge generated through our research programme can be translated into actionable policy advice and public health products.
Over the next decade, we will strengthen these ties and will specifically seek to build partnerships to translate our research insights into products that are sensitive to the needs of the low- and middle-income countries’ populations that are most at risk from the diseases that we study. For example, by developing probiotics, prebiotics, or microbiome-modulating foods that are tailored to local diets and demography, we will aim to maximise improvements in population health.