
Dr Kenny Roberts
Senior Staff Scientist
My research combines state-of-the-art single molecule in situ hybridisation and sequencing technologies with cutting edge high-throughput microscopy to visualise the expression of genes within human tissues in a spatially resolved manner.
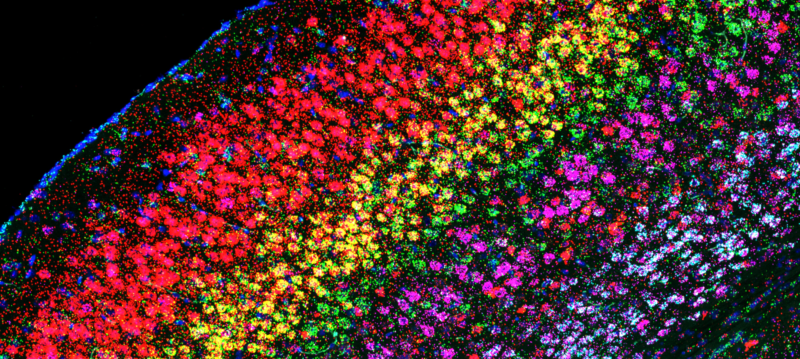
 The world of spatial genomics and transcriptomics has exploded in recent years, along with the breadth of technologies available. I work closely between external collaborators and commercial partners, and Sanger faculty teams and pipelines in order to develop and establish spatial technologies such as Xenium in situ sequencing as resources for the institute. I collaborate with colleagues in histology, microscopy, and image analysis to generate spatially-resolved biological data that complement single-cell transcriptomic approaches.
The world of spatial genomics and transcriptomics has exploded in recent years, along with the breadth of technologies available. I work closely between external collaborators and commercial partners, and Sanger faculty teams and pipelines in order to develop and establish spatial technologies such as Xenium in situ sequencing as resources for the institute. I collaborate with colleagues in histology, microscopy, and image analysis to generate spatially-resolved biological data that complement single-cell transcriptomic approaches.
I am particularly interested in the application of these cutting-edge technologies to biological models and questions. I have previously collaborated with faculty teams in Cellular Genetics and CASM to generate multi-modal atlases of human tissues, including the uterus and placenta (Vento-Tormo), gastrointestinal tract and heart (Teichmann), spine (Behjati), and breast (Yates). I am currently working on diversifying the applications of imaging-based spatial transcriptomics to cultured cells and less common tissue sample formats in order to make these technologies as widely applicable as possible
My timeline
Senior Staff Scientist, High Throughput Spatial Genomics, Cellular Research and Gene Editing R&D
Senior Staff Scientist, High Throughput Spatial Genomics, Cellular Genetics
Staff Scientist, High Throughput Spatial Genomics Initiative
Post-Doctoral Fellow, Wellcome Sanger Institute
Completed D.Phil. Human Genetics and Physiology at the University of Oxford
Awarded M.Biochem., Molecular and Cellular Biochemistry, University of Oxford