The contribution of local animal populations to human drug resistant Salmonella infections may previously have been overstated
A new study has shown that, contrary to popular belief, local domestic animals are unlikely to be the major source of antibiotic-resistant Salmonella in humans. The result comes from a detailed study of DNA from more than 370 Salmonella samples collected over a 22-year period.
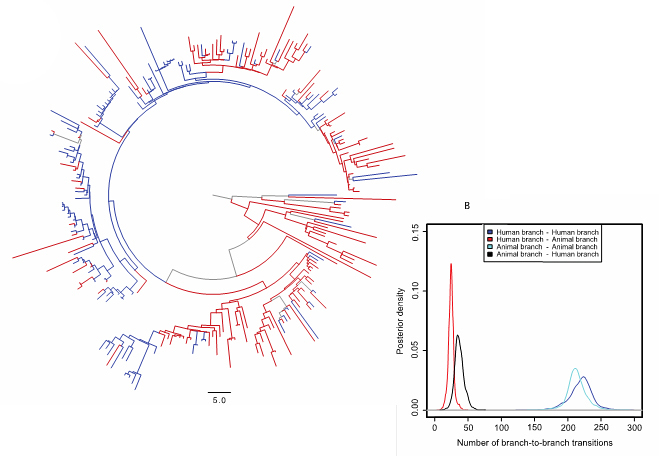
By studying the genetic variation in the Salmonella bacteria and their drug resistance genes, researchers found that distinguishable bacterial populations exist in human and animal populations living side by side. Antibiotic resistance is considered to be one of the most important dangers to human health, threatening to make many treatments to common infections ineffective. By comparing the genomes of Salmonella in humans and animals the researchers have provided important new insights into the likely sources and spread of antibiotic resistant infections. First, the Salmonella bacteria largely remained within their original host populations and second, there were more varied combinations of drug resistance in the human-infecting bacteria.
Salmonella infection is a global issue, with approximately 94 million people contracting gastroenteritis or food poisoning each year. The combined annual cost in the United States and European Union is estimated to be more than £4 billion ($6 billion). This public health issue is exacerbated further by antibiotic resistance, which can lead to more complicated and protracted illness in patients and increased treatment costs.
“For the first time we’ve determined in detail and on a large scale how Salmonella strains taken from humans and animals in the same setting and over the same time period relate to each other. Our genomic data reveal how the Salmonella bacteria spread during the course of a long-term epidemic. We found that people have a more diverse source of infection and antibiotic resistance than just the local animals, pointing towards alternative sources.”
Dr Alison Mather First author on the study, from the Wellcome Trust Sanger Institute
The team sequenced DNA from 373 samples from humans and animals infected with Salmonella Typhimurium DT104 over a 22-year period, mainly from Scotland, but also from other countries. This is the largest study of its type; whole genome DNA sequencing delivers the highest level of resolution possible to examine how closely related the bacteria are, enabling the team to unravel the details of this epidemic.
The team discovered that, contrary to much current thinking, the populations of Salmonella in humans and animals were distinguishable. They also found that the estimated number of times that the bacteria had jumped from animals to humans (and vice versa) was remarkably low. In addition, there was greater diversity in antibiotic resistance genes in salmonellae isolated from humans. Taken together, these findings suggest that the contribution of local animal populations to human infections with S. Typhimurium DT104 may previously have been overstated.
“This is a study that uses the latest genomic approaches and a unique collection of samples to address a significant public health problem. Our data provide a very simple message, challenging the established view that local animals are the predominant source of Salmonella infections in Scotland. This finding will reinvigorate discussions on the sources of antibiotic-resistant Salmonella infections in humans in other environments.”
Professor Nicholas Thomson Senior author from the Wellcome Trust Sanger Institute
The team speculate that international travel and imported foods may be major sources of antibiotic-resistant strains of Salmonella. However, to understand fully the routes of infection and find ways to prevent it, further research into other bacteria and other environments will be needed.
“Discovering that the animal and human populations of Salmonella were as distinguishable as they were was a great surprise to us. This finding in no way undermines the importance of prudent antimicrobial use in all species. But our study does demonstrate that greater effort needs to be focused on understanding the natural history of the pathogens and on identifying the major sources of resistance in our global ecosystems.”
Professor Stuart Reid Co-author from the Royal Veterinary College
More information
Funding
Funding for the research came from, the William Stewart Fellowship at the University of Glasgow, Wellcome Trust grant 098051, funding from the European Union Seventh Framework Programme [FP7/2007-2013] under ERC Grant agreement no. 260864, National Institutes of Health R01 grants AI107034 and HG006139 and National Science Foundation grant DMS-1264153.
Participating Centres
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK.
- Royal Veterinary College, North Mymms, Hatfield, UK.
- Department of Veterinary Medicine, University of Cambridge, Cambridge, UK.
- Scottish Salmonella Shigella and Clostridium difficile Reference Laboratory, Stobhill Hospital, Glasgow, UK.
- National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada.
- Animal Health and Veterinary Laboratories Agency, Weybridge, UK.
- Gastrointestinal Bacteria Reference Unit, Public Health England, Colindale, London, UK.
- Pathogen Genomics Center, National Institute of Infectious Diseases, Tokyo, Japan.
- Bacterial and Parasitic Disease Research Division, National Institute of Animal Health, Ibaraki,Japan.
- Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan.
- Departments of Biomathematics and Human Genetics, David Geffen School of Medicine at UCLA, and Department of Biostatistics, UCLA Fielding School of Public Health, University of California, Los Angeles, USA.
- Department of Microbiology and Immunology, Rega Institute, KU Leuven, Leuven, Belgium.
- Boyd Orr Centre for Population and Ecosystem Health, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, UK.
Publications:
Selected websites
Health Protection Scotland
Health Protection Scotland (HPS) plan and deliver specialist national services which co-ordinate, strengthen and support activities aimed at protecting all the people of Scotland from infectious and environmental hazards. We do this by providing advice, support and information to health professionals, national and local government, the general public and a number of other bodies that play a part in protecting health. HPS also has responsibility for the commissioning of reference laboratory services for NHSScotland.
HPS is part of NHS National Services Scotland, who supports Scotland’s health by delivering shared services and expertise that help other organisations to work more efficiently. In June 2013, HPS was brought together with the Information Services Division to form a new unit, Public Health and Intelligence, within NHS National Services Scotland.
Scottish Salmonella, Shigella and Clostridium difficile Reference Laboratory
The Scottish Salmonella, Shigella and Clostridium difficile Reference Laboratory (SSSCdRL) provides a national reference service for the identification and typing of pathogenic enteric bacteria for the Health Service, public health, veterinary and other agencies in Scotland. As well as providing a source of information and advice, the Laboratory actively participates in training, development and relevant externally-funded research. There is close collaboration with a number of agencies including the Laboratory of Gastrointestinal Pathogens (LGP), London and other international reference centres. The laboratory is a participant in the EC-funded programme organised by the European Centre for Disease Prevention and Control for surveillance of gastrointestinal infection. The SSSCdRL is supported by Health Protection Scotland.
The Royal Veterinary College
Founded in 1791, The Royal Veterinary College is the oldest and largest veterinary school in the English speaking world and a constituent College of the University of London. An independent, research led organisation, it provides support for the veterinary and related professions through its three referral hospitals, diagnostic services and continuing professional development courses.
The Animal Health and Veterinary Laboratories Agency
The Animal Health and Veterinary Laboratories Agency is an executive agency of the Department for Environment, Food and Rural Affairs, working across Great Britain on behalf of Defra, Scottish Government and Welsh Government.
The University of Cambridge
The University of Cambridge’s mission is to contribute to society through the pursuit of education, learning and research at the highest international levels of excellence. It is made up of 31 colleges and more than 100 departments that cater for some 12,000 undergraduate and 6,000 postgraduate students. The University’s reputation for outstanding academic achievement is known worldwide and reflects the intellectual achievement of its students, as well as the world-class original research carried out by the staff of the University and the Colleges. Cambridge is consistently ranked among the top five universities in the world.
Institute of Biodiversity, Animal Health and Comparative Medicine in the College of Medical Veterinary and Life Sciences
The Institute of Biodiversity, Animal Health and Comparative Medicine in the College of Medical Veterinary and Life Sciences at the University of Glasgow is a multidisciplinary Institute that integrates Glasgow’s substantial research expertise in animal biology and ecology with that in comparative and veterinary medicine. Research spans studies from the genomic and cellular levels to individuals, populations, species and ecosystems. This is a unique Research Institute within the UK that links research on animal diseases, production and welfare with ecological and evolutionary approaches. It is driven by the need to create multidisciplinary teams to address some of the major research challenges in relation to environmental change, emerging diseases and the conservation of biodiversity. This Institute meets national and global agendas linking environmental change, food security, infectious diseases, and animal and ecosystem health.
KU Leuven
KU Leuven (University of Leuven) is a leading European research university dedicated to excellent research, education and service to society. It is a founding member of the League of European Research Universities and has a strong European and international orientation. Its sizeable academic staff conducts basic and applied research in a comprehensive range of disciplines. University Hospitals Leuven, its network of research hospitals, provides high-quality healthcare and develops new therapeutic and diagnostic insights with an emphasis on translational research. The university welcomes more than 40,000 students, of which 15.5% are international from more than 140 countries. Its doctoral schools organise internationally oriented PhD programmes for over 4,000 doctoral students.
The National Institute of Infectious Diseases
The National Institute of Infectious Diseases aims at carrying out extensive and original research projects on a variety of contagious diseases from the standpoint of preventive medicine, improving human health and welfare by suppressing infectious diseases, and clarifying and supporting the scientific background of health and medical administration of the government.
Public Health England
Public Health England’s mission is to protect and improve the nation’s health and to address inequalities through working with national and local government, the NHS, industry and the voluntary and community sector. PHE is an operationally autonomous executive agency of the Department of Health.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


