Shigella flexneri persists in environment and switches serotypes
The largest global genomic study of Shigella flexneri, the leading cause of this disease, shows that clear lineages of the bacteria persist within the local environment over many years, a likely clue to the long term success of this pathogen in causing dysentery. This finding, coupled with discovery that the lineages can switch serotype and thereby evade serotype-based vaccines, highlights the importance of achieving the United Nation’s Millennium Development Goals for clean water.
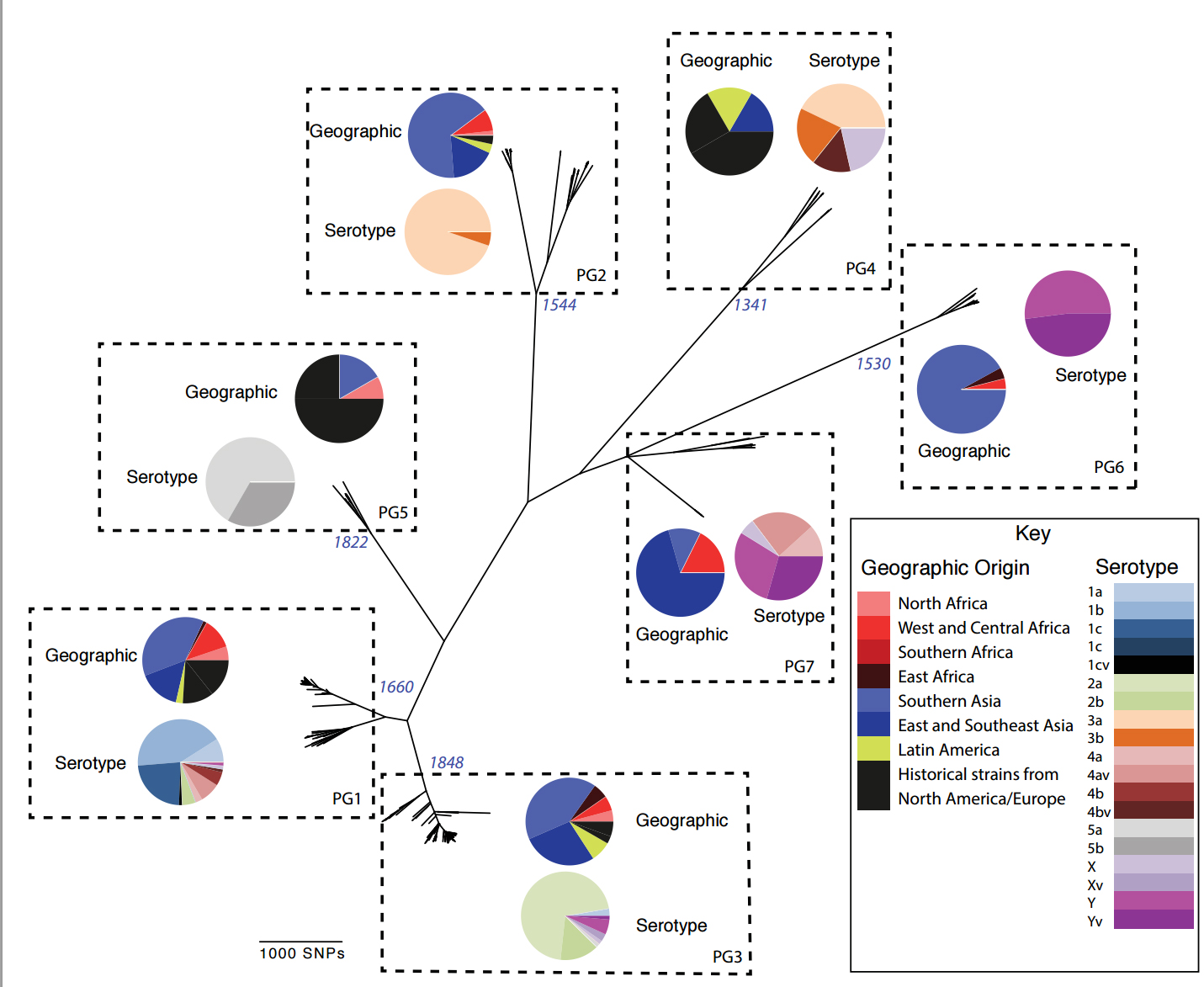
Until now understanding of the spread and evolution of S. flexneri has been based on serotyping, a technology developed more than 50 years ago. To produce greater insight into the bacteria’s evolution over time and reconstruct its worldwide transmission, researchers sequenced the whole genomes of 351 S. flexneri samples from Africa, Asia and South and Central America, and historical collections dating back to 1913.
“Understanding how S. flexneri has changed and spread in endemic countries is vital for developing and targeting interventions more effectively. By using genomics we have been able to unambiguously characterise this pathogen on a global scale and our findings redefine what we knew about this bacteria.”
Thomas Connor Senior first author from Cardiff University School of Biosciences
Analysis revealed distinct groups of S. flexneri that have remained endemic in particular locations and environments for tens or hundreds of years. This finding suggests that the bacteria are not reliant on humans for survival, but can remain viable in the local environment; from which they then re-emerge to cause disease.
“Unlike other Shigella species, we have found that S. flexneri is able to survive in a geographic region, independent of human contact. This tells us that eradicating the bacteria in people through vaccination alone, although important, will not be enough. S. flexneri persists in water for long periods so improving water quality and sanitation, as outlined in the Millennium Development Goals, will be key to preventing reinfection and removing the disease from an area.”
Clare Barker One of the study’s joint first authors from Cardiff University School of Biosciences
The power of genomic analysis has also revealed that using classical microbiological techniques such as serotyping to understand the spread and diversity of the bacteria is unhelpful for planning public health campaigns and for creating effective vaccines. However, the insights produced from sequencing the whole genome offer new options for more targeted vaccine production.
“Our findings show that major lineages of S. flexner are able to switch between serotypes and thereby evade the protective effect of serotype-based vaccination approaches. By using genome sequencing to study the species at the highest resolution possible, we are able to identify clear lineages of bacteria based on the virulence genes they carry. These lineages can then be targeted more effectively for intervention whether that be through vaccine development and/or alternative strategies.”
Nick Thomson Senior last author and member of Faculty at the Sanger Institute


