New malaria drug targets identified in liver stage of life cycle
New research into the different life stages of malaria parasites has identified promising areas for new drug targets aimed at disrupting the parasite’s invasion of the liver. Scientists from the Wellcome Sanger Institute, the University of Bern and Umeå University discovered seven metabolic pathways that the parasite needs to infect the liver, where the parasite multiplies rapidly before invading the blood and causing malaria.
The study, published today in Cell, is the first systematic study of gene function in the Plasmodium malaria parasite family. By ‘deleting’ certain genes from the parasite’s DNA and observing the effect on its behaviour, the team were able to identify which ones are essential for the liver stage of its life cycle. This information will help to develop drugs that target these genes and interrupt the parasite’s ability to cause malaria.
Malaria remains a major global health problem. Between 2000 and 2015, an ongoing drive to eliminate the disease has seen worldwide malaria deaths halve from 864,000 to 429,000 per year.* But the increase in drug-resistant strains of malaria means this progress may be at risk if new forms of treatment are not developed.
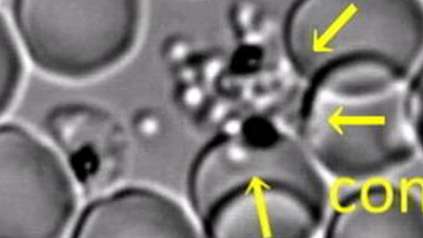
The various species of Plasmodium parasite have complex life cycles that begin in mosquitos before continuing in mammalian hosts, such as humans or rodents. Around 100 parasites transfer to the mammal host through a mosquito bite, where they then move to the liver, a metabolically rich environment that acts as an incubator for them to reproduce rapidly. After 7-10 days, around 10,000 parasites leave the liver to invade red blood cells, where they cause the symptoms of malaria.
In the new study, researchers carried out a genome-wide gene deletion study on the parasite Plasmodium berghei, which infects certain species of rodent, using a malaria mouse model from the Institute of Cell Biology at the University of Bern.
More than a thousand parasite genes were switched off, or ‘knocked out’, with each gene assigned a unique DNA barcode**. The team then used next generation genome sequencing technology at the Wellcome Sanger Institute to count those barcodes and analyse how the removal of individual genes affected the parasite’s life cycle.
The team found 461 genes that are essential for efficient parasite transmission to mosquitoes and through the liver stage back into the bloodstream of mice. From this data, a model of P. berghei liver-stage metabolism was created by scientists in Lausanne that allowed the researchers to pinpoint seven metabolic pathways essential to the parasite’s ability to grow rapidly in the liver.
“To identify seven metabolic pathways that are essential to a Plasmodium parasite’s ability to reproduce in the host liver is incredibly exciting. Our findings will allow malaria researchers worldwide to focus on these essential genes, in order to develop efficient drugs and vaccines to help tackle malaria.”
Dr Ellen Bushell From Umeå University and an alumnus of the Wellcome Sanger Institute
Most anti-malarial drugs target the blood stage of the parasite’s life cycle, but very few target the liver stage. Growing resistance to blood-stage malaria drugs, such as artemisinin, makes the possibility of new liver-stage drugs increasingly important.
A further benefit to drugs that target Plasmodium parasites in the liver would be their effectiveness against forms of malaria, such as Plasmodium vivax, that can lay dormant in the liver and cause recurrence of symptoms for years after the initial infection.
“The world has achieved great success in fighting malaria by targeting the blood stage of Plasmodium parasites, halving the number of malaria deaths in less than two decades. But Plasmodium parasites have repeatedly and rapidly developed resistance to all available blood-stage drugs. Liver-stage parasites are an important reservoir of infection, but because there are far fewer of them it makes the development of drug resistance at this stage less likely. The discovery of new liver-stage drug targets is therefore both timely and important.”
Dr Oliver Billker From Umeå University and an alumnus of the Wellcome Sanger Institute
The research approach was only possible through a combination of the vast sequencing and cloning capacities at the Wellcome Sanger Institute and the extraordinary infrastructure at the Institute of Cell Biology in Bern, where the life cycle of the malaria parasite was established.
“Twenty-two international scientists from the fields of molecular biology, parasitology, statistics and mathematical modelling participated in this project. This illustrates the effort in conducting this study, analysing the data and modelling the experimental findings to bring them into a meaningful context.”
Professor Volker Heussler From the Institute of Cell Biology, University of Bern
More information
*For more information on these figures and the state of malaria in general, see the WHO World Malaria Report 2016 https://www.who.int/malaria/media/world-malaria-report-2016/en/
**More information about the life cycle of Plasmodium parasites and malaria is available at the YourGenome website: https://www.yourgenome.org/video/malaria-an-introduction
***The DNA Barcoding method involves tagging specific genes with molecular barcodes. These barcodes enable scientists to identify individual mutants and measure the growth of the genetically modified parasite by counting the number of barcodes using a benchtop sequencer. For more information see: Ana Rita Gomes et al. (2015). A genome scale vector resource enables high throughput reverse genetic screening in a malaria parasite. Cell Host Microbe 17, 1–10.
Publication:
Rebecca R. Stanway et al. (2019). Genome Scale Identification of Essential Metabolic Processes for Targeting the Plasmodium Liver Stage. Cell. DOI: 10.1016/j.cell.2019.10.030
Funding:
This study was funded by a MalarX grant (2013/155) and a SystemsX grant awarded by the SNSF. Work conducted at the Wellcome Sanger Institute was funded by the core Wellcome grant (206194/Z/17/Z).
Selected websites
The University of Bern
The University of Bern is a public institution, based in Switzerland’s capital city, with a strong focus on providing excellence in research and teaching. The university has some 18,400 students benefiting from a full range of courses in most disciplines at undergraduate and graduate levels, as well as continuing education. The University of Bern is one of the world’s leading universities in climate research, space research as well as health and medicine. It hosts international research platforms, national centers of competence in research and trans- and interdisciplinary programs. The institution offers state-of-the-art facilities to its students and teaching staff at its eight faculties. http://www.unibe.ch/index_eng.html
Umeå University
Umeå University is one of Sweden’s largest institutions of higher learning with over 33,000 students and 4,000 employees. We have a well-established international research profile and a broad range of study options. Our campus constitutes an inspiring environment that encourages interdisciplinary meetings – between students, researchers, teachers and external stakeholders. Through collaboration with other members of society, we contribute to the development and strengthen the quality of our research and education. https://www.umu.se/en/
Wellcome Sanger Institute
The Wellcome Sanger Institute is a world-leading genomics research centre. We undertake large-scale research that forms the foundations of knowledge in biology and medicine. We are open and collaborative; our data, results, tools and technologies are shared across the globe to advance science. Our ambition is vast – we take on projects that are not possible anywhere else. We use the power of genome sequencing to understand and harness the information in DNA. Funded by Wellcome, we have the freedom and support to push the boundaries of genomics. Our findings are used to improve health and to understand life on Earth. Find out more at scion-02.sandbox.sanger.ac.uk or follow us on Twitter, Facebook, LinkedIn and on our Blog.
About Wellcome
Wellcome exists to improve health by helping great ideas to thrive. We support researchers, we take on big health challenges, we campaign for better science, and we help everyone get involved with science and health research. We are a politically and financially independent foundation. https://wellcome.org/