New insights into the evolutionary history of gorillas
Scientists have uncovered evidence for gene flow from an extinct gorilla population to eastern gorillas, providing new insights into the evolutionary history of our relatives and ancestors. They suggest ancient interbreeding events left a genetic legacy, detectable in the genomes of present-day eastern gorillas. The discovery mirrors how modern humans and bonobos similarly carry genes from ancient, extinct groups in their DNA.
These interbreeding events with a ‘ghost’ gorilla population were estimated to have occurred around 40,000 years ago, before the split between mountain and lowland eastern gorillas. A ‘ghost’ population refers to refers to an ancient or extinct group of organisms with no fossil record – only genetic traces in the genomes of present-day individuals that point to their existence.
Researchers from the University of Vienna, the Institute of Evolutionary Biology in Barcelona and their collaborators from the Wellcome Sanger Institute analysed the genomes of gorillas using modern statistical methods, including neural networks*. They found approximately 3 per cent of the genome of these eastern gorillas consists of a DNA contribution due to interbreeding. Western gorillas do not share this feature.
Further, the ancient interbreeding may have had an impact on the ability of eastern gorillas to perceive bitter tastes, offering a unique window into the interaction between the genetic makeup and behaviour of eastern gorillas today.
The findings, published in Nature Ecology and Evolution (27 July 2023), highlight the genetic diversity in modern gorilla populations and help piece together the unknowns about how this species has interacted and evolved over time.
Throughout human evolution, there have been several instances where ancient human-like relatives, Neanderthals and Denisovans, interbred with modern humans, leaving a lasting impact on our genetic makeup. Similar interbreeding has also been identified in chimpanzees and bonobos, two other great ape species closely related to humans.
In this new study, scientists set out to investigate whether such interbreeding also occurred in gorillas, in order to better understand their evolutionary history.
Gorillas are composed of two species – western and eastern gorillas, each of which has two subspecies: western gorillas include the western lowland gorillas and the cross river gorillas, while eastern gorillas include the eastern lowland gorillas and the closely related mountain gorillas.
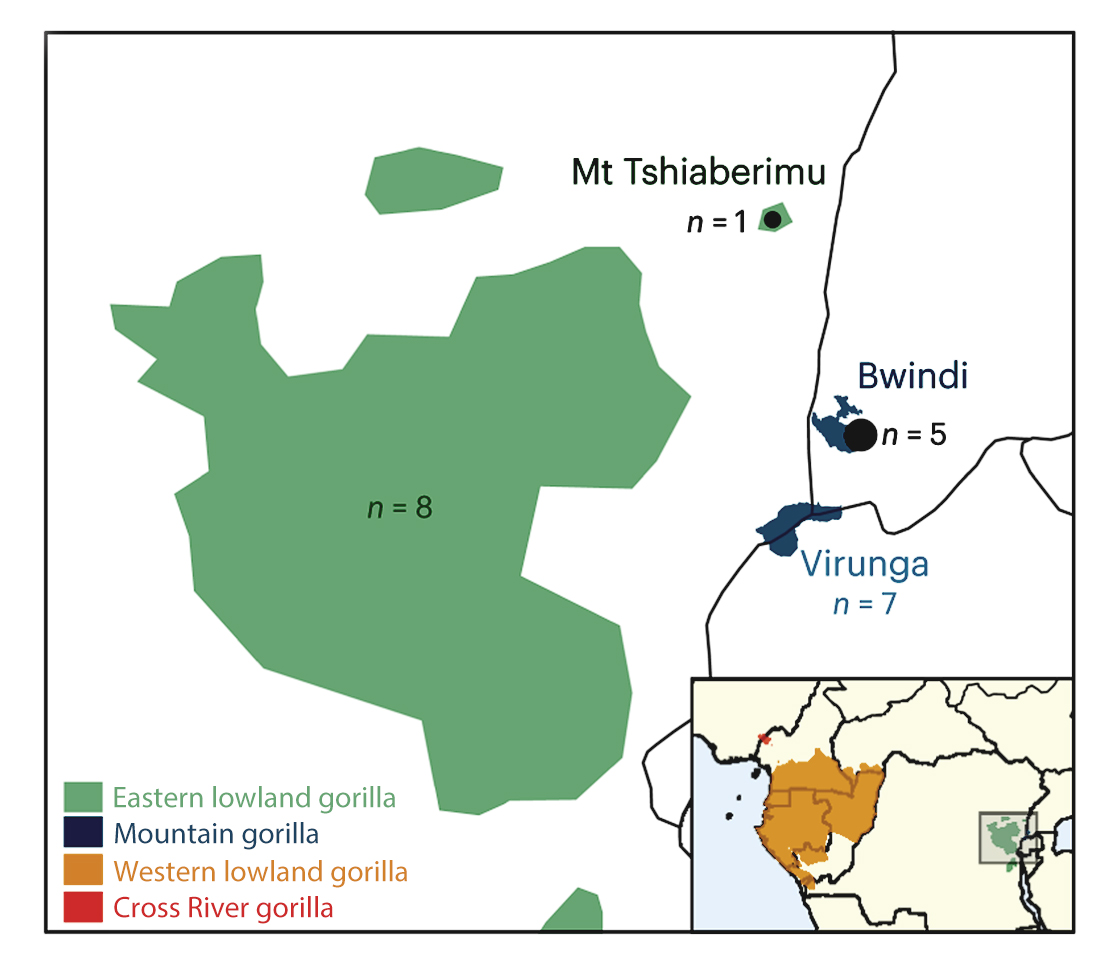
Researchers at the Institute of Evolutionary Biology in Barcelona analysed whole genomes of all four gorilla subspecies using sophisticated statistical methods** and neural networks. They were able to include blood samples from six individuals from previously unexplored regions, including Bwindi National Park in Uganda, one of only two places where the few remaining mountain gorillas can be found. The Wellcome Sanger Institute received and whole-genome sequenced the new gorilla samples.
The international team discovered that 40,000 years ago, genes were exchanged between a now-extinct gorilla ‘ghost’ population and the common ancestor of the eastern lowland gorillas and the mountain gorillas.
Up to 3 per cent of the genome of today’s eastern gorillas includes remains of genes from this ‘ghost’ population, which separated from the common ancestors of all gorillas more than 3 million years ago.
The research sheds new light on how ancient genes may contribute to evolution but also shows how genes from long-extinct ancestors can still have important functions in the lives of animals living today***. For example, the researchers found that a gene responsible for taste, particularly the ability to detect bitterness, was introduced from the ghost population into today’s eastern lowland gorillas and mountain gorillas. This gene might have been useful for these gorillas because it helps them avoid eating poisonous or bitter-tasting food, making it an essential survival trait.
“Genome sequences provide powerful insights into the history of a species. They previously told us that a million years ago was a good time to be a gorilla, with large thriving populations, and that numbers have declined steadily since then leading to the tiny endangered present-day populations. The new work shows some of the unsuspected twists in evolutionarily more recent times, just 40,000 years ago, and makes us wonder whether we should now expect to find complex mixing of distant populations in the history of primates more generally.”
Chris Tyler-Smith, author and retired Senior Group Leader at the Wellcome Sanger Institute
“Our study gives us a better insight into the evolutionary history of gorillas and provides a valuable contribution to help us better understand which effects gene flows from extinct populations can have on current populations.”
Tomas Marques-Bonet, senior author of the study and professor of genetics at the Department of Medicine and Life Sciences (MELIS) at the Institute of Evolutionary Biology in Barcelona (UPF)
More information
This work was supported by the Uganda Wildlife Authority for the Gorilla monitoring and research permission.
*Neural networks are used to simplify and analyse the data used in the study. They help to take into account any differences between observed and simulation-generated data to better understand the relationships and patterns in the data. This allows scientists to draw more accurate conclusions from their analysis.
**The researchers used an approximate Bayesian computation (ABC) approach based on a previous implementation in the Pan clade to find evidence for introgression from an extinct lineage into the common ancestor of eastern gorillas. For the full methods, please refer to the publication.
***Another example, this time likely impacting gene function negatively, were the remains from genes from the ancient ‘ghost’ population on their X chromosome, or rather their surprising scarcity. The chromosome is particularly subject to negative (or purifying) selection – the body’s ‘quality control’ mechanism – which means harmful genetic changes tend to be eliminated over time, preventing them being passed on in a population. One possible reason for this could be that the X chromosome exists in only one copy in male individuals, unlike other chromosomes. Therefore, harmful mutations may have a stronger effect.
Publication:
- Pawar, C. Tyler-Smith, Y. Xue, J. Prado-Martinez, T. Marques-Bonet, M. Kuhlwilm et al. (2023) Ghost mixture in eastern gorillas. Nature Ecology and Evolution. DOI: 10.1038/s41559-023-02145-2
Funding:
This research was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme and the Vienna Science and Technology Fund (WWTF) and by Wellcome. For full funding acknowledgements, please refer to the publication.


