Genomic surveillance of antibiotic resistance in the Philippines established
Antibiotic resistance surveillance in the Philippines has moved into the genomic era, enabling better tracking of dangerous bacteria. Researchers at the Centre for Genomic Pathogen Surveillance (CGPS housed at the Wellcome Sanger Institute and The Big Data Institute (BDI), University of Oxford), and the Philippine Research Institute for Tropical Medicine (RITM), set up local DNA sequencing and analysis of drug resistant bacteria in the Philippines. This genomic capacity has enhanced ongoing national infection control including tracking the spread of resistance to last-line antibiotics and identifying drug resistant infections in a hospital baby unit, helping control the outbreak.
Reported in Nature Communications this week, this study shows the power of local genomic sequencing within national surveillance networks in low- and middle-income countries, and could be extended to other locations to tackle the global challenge of antimicrobial resistance.
Antimicrobial resistance (AMR) is a global health problem, with resistance to common antibiotics found in all regions of the world. This means it can be extremely difficult to treat some bacterial diseases such as MRSA, tuberculosis and gonorrhoea, and raises risks of any surgery.
Surveillance of AMR is critical to understand and try to halt its spread, and DNA sequencing can pinpoint resistance mechanisms and uncover transmission patterns. However, genomic surveillance is less common in low- and middle-income countries (LMICs), which are predicted to be the most affected by AMR.
The Philippines has a very well established Antimicrobial Resistance Surveillance Programme within the Philippine Department of Health, which uses laboratory-based methods to track antimicrobial resistance. In 2018 the researchers helped set up a DNA sequencing facility within this to build local capacity for genomic surveillance in the Philippines*. This has included establishing local capacity in genomics and data interpretation through shared training.
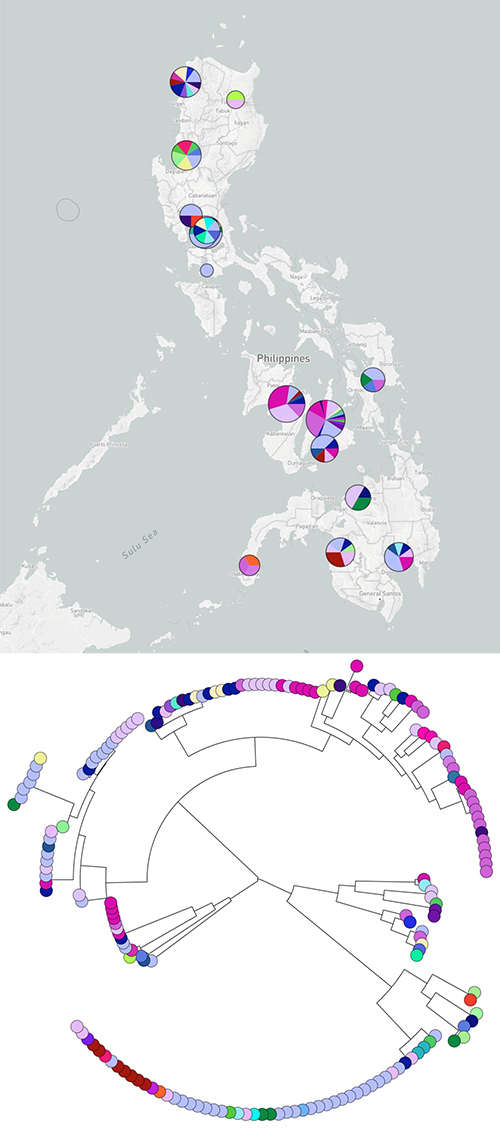
Samples were sequenced from more than 20 sites across the Philippines, focusing on bacteria that are resistant to the last-line antibiotics, and listed by the World Health Organisation as top priority pathogens for the development of new antibiotics**. The teams collectively analysed the data, creating phylogenetic trees that showed how the bacterial strains are related to each other, and uncovered several high-risk clones.
Combining the genetic findings with epidemiological data allowed the researchers to pinpoint strains in particular locations. In one hospital they identified a cluster of the same strain of carbapenem-resistant Klebsiella pneumoniae in a neonatal intensive care unit, and revealed that this was being spread within the hospital. This evidence enabled the hospital to bolster their infection control team, to control potential future outbreaks.
“Here in the Philippines we have more than 30 years of experience developing laboratory methods to track AMR, with our Antimicrobial Resistance Surveillance Program. Now, working with our partners in the UK, we have established local capacity and expertise for whole genome sequencing in the Philippines, adding genomic surveillance to these other methods. This is helping us to identify emerging resistant strains much faster, so we can understand what is happening, prevent transmission of AMR and save lives.”
Dr Celia Carlos, joint lead of the project from the Research Institute for Tropical Medicine, Philippines
“The programme not only helped set up the genomic infrastructure in the Philippines, but also enabled close collaboration between the teams in the UK and the Philippines. This included exchange visits between the researchers and training to transfer ownership of the sequencing, analysis and understanding to the team in the Philippines, and ensured that everyone understood the resourcefulness and challenges of the sentinel sites.”
Dr Silvia Argimón, first author on the paper from the Centre for Genomic Pathogen Surveillance
Genomic surveillance allows the team to describe drug-resistant bacteria in terms of their strains, which genes enable the resistance, and how those genes are transferred between bacteria. Through genomics the Philippines now have a greater lens on AMR at the local, the national and international scale, allowing data analysis at a previously difficult level. The data are shared with Philippine public health agencies and with the WHO to inform both local and global understanding of the spread of carbapenam resistance.
“Understanding national dynamics in antimicrobial resistance is important in every country in the world to prevent spread globally, and new technology and tools that enhance this capacity are required. The work by the Philippines team to establish genomics within a national surveillance network is an exemplar for adoption that could be extended to tackle the global challenge of antimicrobial resistance or other infections.”
Professor David Aanensen, Director of the Centre for Genomic Pathogen Surveillance and joint lead on the project
More information
* Whole genome sequencing started at the Antimicrobial Resistance Surveillance Research Laboratory in 2018 with the Illumina MiSeq equipment available locally. A new dedicated bioinformatics server was installed there for sequence data storage and analysis. Data sharing via interactive web tools such as Microreact (www.microreact.org) and Pathogenwatch (www.pathogen.watch) helped the collective data interpretation.
** WHO List of bacteria for which new antibiotics are urgently needed.
https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
Publication:
Silvia Argimón et al. (2020) Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nature Communications. DOI: s41467-020-16322-5
Funding:
This work was funded by the Newton Fund, Medical Research Council (UK) grant MR/N019296/1, Philippine Council for Health Research and Development project number FP160007, National Institutes of Health (NIH), National Institute for Health Research (UK) Global Health Research Unit on genomic Surveillance of AMR (16_136_111) and by the Centre for Genomic Pathogen Surveillance.
Selected websites
Research Institute for Tropical Medicine (RITM)
Established in 1981, the Research Institute for Tropical Medicine (RITM) is the research arm of the Philippine Department of Health (DOH). It is home to 12 National Reference Laboratories (NRLs) for various infectious diseases that provide technical expertise in the laboratory diagnosis and characterization of infectious agents in the country for surveillance, outbreak investigations, and research. RITM also conducts clinical trials and provides clinical care to people suffering from emerging and re-emerging infectious and tropical diseases.
Centre for Genomic Pathogen Surveillance
The Centre for Genomic Pathogen Surveillance is an initiative based at The Big Data Institute at the University of Oxford and The Wellcome Genome Campus focussed on genomic epidemiology, laboratory and software engineering for global surveillance of microbial pathogens. The Centre seeks to provide genomic and epidemiological big data and tools to allow researchers, doctors and governments worldwide to track and analyse the spread of pathogens and antimicrobial resistance.
The NIHR Global Health Research Unit of Genomic Surveillance of Antimicrobial Resistance
The NIHR Global Health Research Unit on Genomic Surveillance of AMR is focussed on capacity building for genomic surveillance in Low and Middle Income Countries. Capacity in Laboratory, Bioinformatics and Financial management within The Phillipines, Nigeria, Colombia and India is driven by National Units linked to networks or participating hospitals and laboratories. Through large scale sequencing, training and delivery of open data through web resources, data are being generated to understand the population structure and monitoring of important species driving the spread of antimicrobial resistance.
The Big Data Institute
The Big Data Institute is located in the Li Ka Shing Centre for Health Informatics and Discovery at the University of Oxford. It is an interdisciplinary research centre that focuses on the analysis of large, complex data sets for research into the causes, consequences, prevention and treatment of disease. Research is conducted in areas such as genomics, population health, infectious disease surveillance and the development of new analytic methods. The Big Data Institute is supported by funding from the Medical Research Council, the UK Research Partnership Investment Fund, the National Institute for Health Research Oxford Biomedical Research Centre, and philanthropic donations from the Li Ka Shing and Robertson Foundations
The Wellcome Sanger Institute
The Wellcome Sanger Institute is a world leading genomics research centre. We undertake large-scale research that forms the foundations of knowledge in biology and medicine. We are open and collaborative; our data, results, tools and technologies are shared across the globe to advance science. Our ambition is vast – we take on projects that are not possible anywhere else. We use the power of genome sequencing to understand and harness the information in DNA. Funded by Wellcome, we have the freedom and support to push the boundaries of genomics. Our findings are used to improve health and to understand life on Earth. Find out more at scion-02.sandbox.sanger.ac.uk or follow us on Twitter, Facebook, LinkedIn and on our Blog.
Wellcome
Wellcome exists to improve health by helping great ideas to thrive. We support researchers, we take on big health challenges, we campaign for better science, and we help everyone get involved with science and health research. We are a politically and financially independent foundation.



