MERS Co-V genomes reveal complex transmission patterns
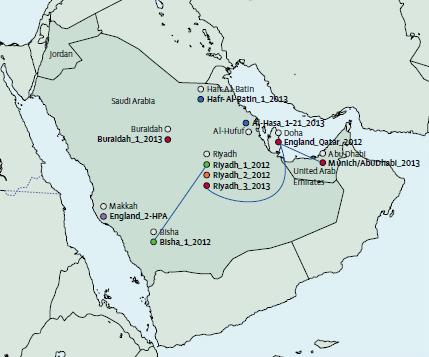
Genome sequencing has identified several infection transmission chains of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in humans. The study published in The Lancet, which produced the largest number of MERS-CoV genomes described to date, provides evidence that MERS-CoV transmission patterns are more complicated than previously considered.
Globally, 111 people have been diagnosed with MERS-CoV since 2012, including 52 deaths. Researchers from the Wellcome Trust Sanger Institute, Edinburgh University and University College London are working with the Kingdom of Saudi Arabia’s Ministry of Health to sequence the virus to discover how it is spreading. This knowledge will help researchers develop interventions and infection-control measures.
Sanger Institute researchers sequenced and analysed the genomes of MERS-CoV samples taken from 21 patients across the Kingdom of Saudi Arabia. The researchers combined the geographic locations of the people with the time they were infected and the amount of genetic differences seen between the virus genomes. This work gave a higher resolution picture of how the virus has spread and how its genome has changed over time.
“We deep-sequenced the genomes of MERS-CoV taken from 21 infected people to calculate accurately the rate of evolution of the virus. Using this evolution rate, we could define genetically plausible transmission pairs. However, of the 13 transmission events that were predicted from the epidemiology, the genetic evidence we gathered could only support eight.”
Dr Matthew Cotten from the Sanger Institute and the study’s first author
The findings suggest that human-to-human transmission is more complicated than expected, and indicates that additional sources of the virus, either human or animal, are involved. One possibility is that there may be undetected (and possibly asymptomatic) people who could be carrying and spreading the virus.
“The genome differences we discovered in some infected people were too great to be explained by replication errors occurring in the virus as it is passed from human to human during a single chain of infection. Instead our findings suggest that different lineages of the virus have originated from the virus jumping across to humans from an animal source a number of times.”
Professor Paul Kellam Senior author from the Sanger Institute
As yet no animal with MERS-CoV has been identified in the Middle East or elsewhere and studies based on small sequence fragments suggest that a common ancestor of the virus may have existed in bats many years ago. Field studies of all the likely reservoir species, including camels, bats, goats, sheep, dogs, cats, rodents and others in the Kingdom of Saudi Arabia and other middle-eastern countries are on-going.
“Further MERS-CoV genomic studies need to be carried out in conjunction with investigations into the recent exposures and activities of infected people. The animal source of MERS-CoV and the way that it is transmitted to humans is not yet known. This information is critical for developing interventions for reducing the risk of transmission, defining the epidemiology and developing effective control measures.”
Corresponding author Professor Ziad Memish Deputy Minister of Health, Riyadh, Kingdom of Saudi Arabia
“Two mass gatherings events attracting over 8 million pilgrims have occurred in Mecca, Saudi Arabia since the discovery of MERS-CoV 12 months ago – the annual Hajj in October 2012, and the recent July 2013 Ramadaan Umrah season – and yet no MERS-CoV cases have been reported from these events to date. In lieu of our study’s genomic findings, watchful surveillance and vigilance is required despite the current minimal risk of global spread.”
Senior Professor Zumla Senior co-author from University College London
More information
Notes to Editors
Funding
The support of all staff at the KSA Ministry of Health and the Jeddah regional laboratory is gratefully acknowledged. The sequencing work was supported by the Wellcome Trust Sanger Institute and the European Community’s Seven Framework Programme (FP7/2007-2013) under the project EMPERIE, European Community grant agreement number 223498 and under the project PREDEMICS, grant agreement number 278433. Professor Zumla (A.Z) acknowledges support from the National Institute of Health Research Biomedical Research Centre, University College London Hospitals, the EDCTP and the EC-FW7.
Participating Centres
A full list of participating centres is available from the paper.
Publications:
Selected websites
UCL
Founded in 1826, UCL was the first English university established after Oxford and Cambridge, the first to admit students regardless of race, class, religion or gender and the first to provide systematic teaching of law, architecture and medicine.
We are among the world’s top universities, as reflected by our performance in a range of international rankings and tables. According to the Thomson Scientific Citation Index, UCL is the second most highly cited European university and the 15th most highly cited in the world.
UCL has nearly 25,000 students from 150 countries and more than 9,000 employees, of whom one third are from outside the UK. The university is based in Bloomsbury in the heart of London, but also has two international campuses – UCL Australia and UCL Qatar. Our annual income is more than £800 million.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


