Putting parasites on the world map
Researchers have developed a new technique to identify hotspots of malaria parasite evolution and track the rise of malarial drug resistance, faster and more efficiently than ever before.
For the first time, researchers have the ability to analyse malaria genomes straight from patient blood samples using new sequencing technologies and informatics methods. As a proof of principle, the team conducted the first analysis of clinical samples from six countries and uncovered unique differences in malaria development in Africa, Asia and Oceania. This study is published in Nature on the 13 June 2012.
Severe forms of malaria infection are caused by the parasite Plasmodium falciparum, which is spread by mosquitoes. Malaria infects over 200 million people and kills approximately 600,000 people every year, primarily children under the age of five in sub-Saharan Africa.
“One of the most striking features of P. falciparum is its ability to evolve, and overcome anti-malarial drugs. Chloroquine has become ineffective against malaria, and resistance to the other frontline drugs is emerging. If we want to control resistance, we first need to be able to monitor the genetic diversity of P. falciparum and identify hotspots of potential resistance as they occur. Rapid sequencing of parasite genomes from the blood of infected people is a powerful way of detecting changes in the parasite population, and potentially an important new surveillance tool in the armamentarium for controlling malaria.”
Professor Dominic Kwiatkowski Senior author of the study from the Wellcome Trust Sanger Institute and Oxford University
The team developed a new technique to extract the parasite DNA directly from blood removing as much human DNA from the sample as possible. The new method overcomes the need to grow the parasite in a blood culture before sequencing, speeding the process and minimising replication errors.
P. falciparum genomes are particularly difficult to sequence because, unlike human DNA, large parts of the DNA sequence are repeated. As a result, the reconstruction of whole parasite genome DNA sequences is slow, expensive and error-prone using current DNA sequencing methods. To avoid these problems, the team used sequence data to create a list of single DNA letter changes, known as SNPs, which can be reliably identified in the gene-rich areas of the genome. These SNPs allow the discovery and measurement of variability in natural parasite populations.
“We catalogued approximately 86,000 SNPs in the parasite genome that allow us to pinpoint differences between parasites around the world, a starting point for understanding how these populations adapt to changes in their environment.”
Dr Magnus Manske Co-first author from the Sanger Institute
Many malaria patients, especially in Africa, are continually infected by malaria parasites, and we have created a new tool for studying the genetic diversity within a single patient, and compare it to the diversity in their environment.”
Dr Olivo Miotto from the Sanger Institute and Oxford University and a co-first author
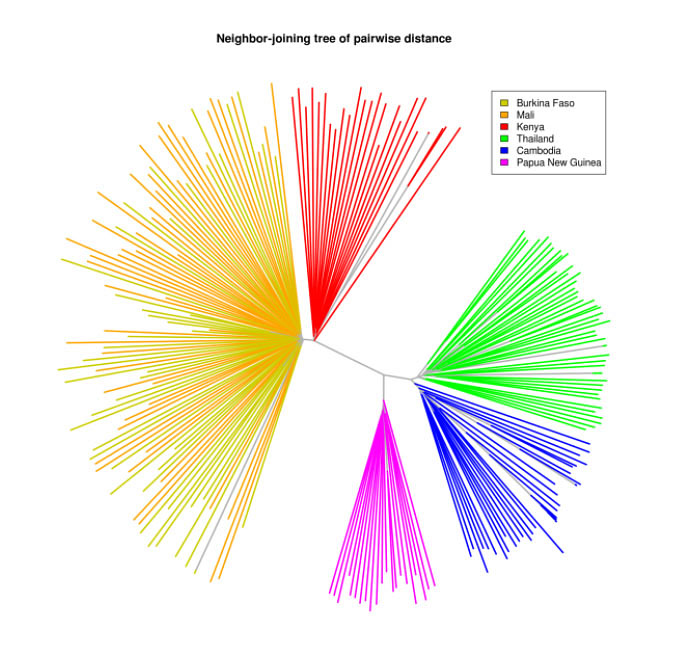
The team used these techniques to analyse samples from Burkina Faso, Cambodia, Kenya, Mali, Papua New Guinea and Thailand. They found that a single infected person could harbour many genetically different malarial parasites, allowing the parasite populations to swap DNA to create new forms. Hence, the pace of parasite evolution is drastically affected by human factors, as well as geography.
Samples taken from people in the neighbouring African countries of Burkina Faso and Mali, where there are very high levels of malaria transmission, showed strong intermingling of P. falciparum genomes.
In stark contrast, Asian P. falciparum parasites collected on the Thai-Burmese border were not only different from those in Africa, but also distinct from those found near the Thai border with Cambodia. This lack of intermingling could be the result of effective malaria control in Thailand, combined with a history of restricted travel of people between Thailand and Cambodia.
“The emergence and spread of anti-malarial drug resistance is a major threat to current global initiatives to control and eliminate malaria. This research provides fundamental insights into the population structure and evolution of Plasmodium falciparum that are essential if we are to identify, map, and then contain spreading resistance. Working as a global community, we can now build on this technique to identify hotspots of antimalarial drug resistance around the world and contain them effectively.”
Professor Nick White of Oxford University and Mahidol University, Thailand
More information
Funding
This research was funded by the Wellcome Trust, the Medical Research Council, Howard Hughes Medical Institute.
Participating Centres
A full list of participating centres can be found in the paper.
Publications:
Selected websites
Oxford University's Medical Sciences Division
Oxford University’s Medical Sciences Division is one of the largest biomedical research centres in Europe, with over 2,500 people involved in research and more than 2,800 students. The University is rated the best in the world for medicine, and it is home to the UK’s top-ranked medical school.
From the genetic and molecular basis of disease to the latest advances in neuroscience, Oxford is at the forefront of medical research. It has one of the largest clinical trial portfolios in the UK and great expertise in taking discoveries from the lab into the clinic. Partnerships with the local NHS Trusts enable patients to benefit from close links between medical research and healthcare delivery.
A great strength of Oxford medicine is its long-standing network of clinical research units in Asia and Africa, enabling world-leading research on the most pressing global health challenges such as malaria, TB, HIV/AIDS and flu. Oxford is also renowned for its large-scale studies which examine the role of factors such as smoking, alcohol and diet on cancer, heart disease and other conditions.
Medical Research Council
For almost 100 years the Medical Research Council has improved the health of people in the UK and around the world by supporting the highest quality science. The MRC invests in world-class scientists. It has produced 29 Nobel Prize winners and sustains a flourishing environment for internationally recognised research. The MRC focuses on making an impact and provides the financial muscle and scientific expertise behind medical breakthroughs, including one of the first antibiotics penicillin, the structure of DNA and the lethal link between smoking and cancer. Today MRC funded scientists tackle research into the major health challenges of the 21st century.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


