
Mutographs of Cancer - CRUK Grand Challenge Project
Summary
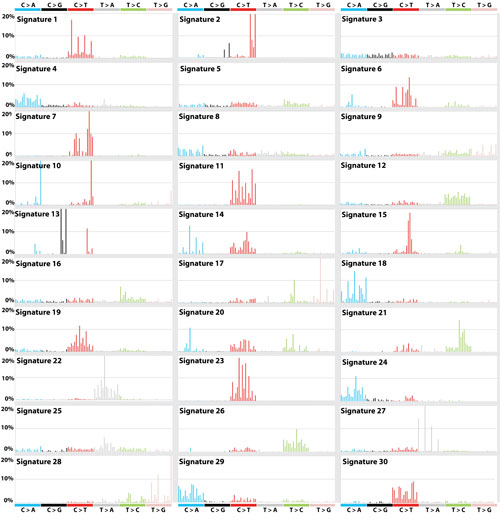
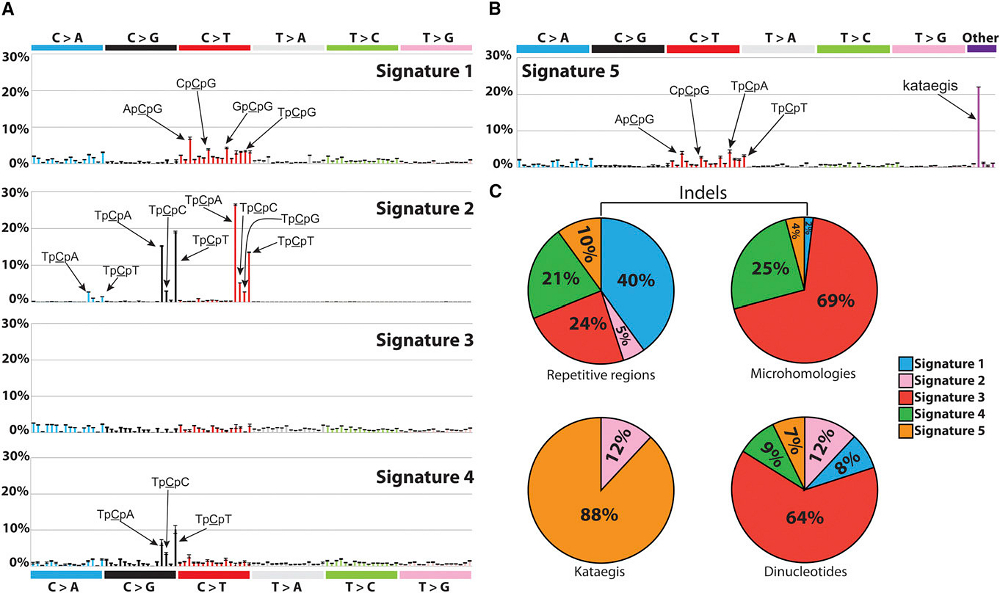
Human behaviours such as smoking and drinking alcohol, and enviromental factors such as UV light can cause cancer by damaging cells’ DNA. This damage occurs in distinctive patterns – known as ‘mutational signatures’ – that are unique to the factor that caused the damage. So far 50 cancer-associated mutational fingerprints have been identified, but only half of them have known cause.
To identify the unknown cancer-causing factors and reveal how they lead to cancer, the genomes of 5,000 pancreatic, kidney, oesophageal and bowel cancer patients from five continents will be studied and their mutational signatures compared. Information about the patients’ about the habits, lifestyles and environments will be gathered to help elucidate causative agents. The researchers will also contine to search for other mutational signatures that haven’t yet been found.
Background
Over the past century, we have learnt much about the environmental, lifestyle, genetic and other factors that cause human cancer. This knowledge has provided the foundation for successful cancer prevention programmes including those aimed at:
- reducing tobacco smoking
- eradicating carcinogenic viruses
- eliminating environmental and industrial exposures such as radiation and asbestos
- reducing breast and ovarian cancer in women with high inherited risk.
However, many common cancers exhibit major differences in incidence between different geographical areas and trends over time for which we do not understand the reasons despite determined investigation. Therefore, important causes of cancer and opportunities for prevention remain to be identified.
All cancers are caused by changes in the DNA within cells in the body that occur over the course of an individual’s lifetime – somatic mutations. Different patterns of somatic mutation, known as “mutational signatures”, are generated by the different environmental, lifestyle and genetic factors that cause cancer.
 For example, tobacco smoke and ultraviolet radiation in sunlight both cause cancer by producing somatic mutations; however, the particular mutational signature caused by tobacco smoke chemicals is found in lung cancers and the distinct mutational signature of ultraviolet light is found in skin cancers.
For example, tobacco smoke and ultraviolet radiation in sunlight both cause cancer by producing somatic mutations; however, the particular mutational signature caused by tobacco smoke chemicals is found in lung cancers and the distinct mutational signature of ultraviolet light is found in skin cancers.
Recently, through analysis of the DNA sequences of many thousands of cancers of diverse types from across the world, approximately 40 different mutational signatures have been reported. However, the environmental, lifestyle, genetic or other potential causes of many of these mutational signatures are unknown.
Overall Goals
The overall goal of this “Mutographs of Cancer” CRUK Grand Challenge Project is to advance understanding of the causes of cancer through studies of mutational signatures. This will be achieved by:
- Investigating whether different mutational signatures in the DNA of cancers explain geographic differences in cancer incidence by collecting cancer samples and information about cancer-causing exposures from 5,000 patients with colorectal, oesophageal, pancreatic and kidney cancer living in regions of high or low cancer incidence in five continents, sequencing the DNA of these cancers and comparing the mutational signatures present.
- Identifying specific causes of mutational signatures by sequencing the DNA of rodent cancers and cultured human cells experimentally exposed to 150 cancer-causing agents, thus assembling a compendium of mutational signatures associated with known causes of cancer.
- Investigating whether mutational signatures in the DNA of normal cells can be used to understand and monitor cancer-causing exposures in healthy people by sequencing the DNA of normal lung, kidney, liver and blood from people who have been exposed to cancer-causing agents.
- Understanding the causes of cancer, our work may lead to new approaches to prevent it and provide opportunities for more effective application of therapies.
Structure of the Research
Through studying the mutational signatures present in the genomes of cancer and normal cells we aim to comprehensively catalogue the mutational processes that cause human cancer, to understand their causes and to apply this knowledge to cancer prevention. To achieve this the research has been structure into work packages to deliver three Specific Aims:
- To elucidate the causes of major global geographical and temporal differences in cancer incidence through mutational signatures.
- To identify, characterise and understand the biological processes underlying mutational signatures.
- To survey and monitor mutagenic exposures in normal cells in humans through mutational signatures.
Specific Aim 1: To elucidate the causes of major global geographical and temporal differences in cancer incidence through study of mutational signatures
- Work Package 1: Mutational signatures in five cancer types across five continents. Led by Paul Brennan, IARC – International Agency for Research on Cancer
- Work Package 2: Development of computational approaches for mutational signature analysis. Led by Ludmil Alexandrov, University of California, San Diego
Specific Aim 2: To identify and characterise the biological processes underlying mutational signatures
- Work Package 3: Mutational signatures of cancer development in rodents exposed to known or suspected carcinogens. Led by Allan Balmain, University of California, San Francisco
- Work Package 4: Establishing a compendium of mutational signatures through exposure of cells in culture to defined mutagens. Led by David Phillips, King’s College London
Specific Aim 3: To survey and monitor mutagenic exposures in normal cells in humans through mutational signature analysis
- Work Package 5: Understanding mutation burdens and mutational signatures in non-cancer tissues. Led by Peter Campbell, Wellcome Sanger Institute
- Work Package 6: Surveying mutagenic exposures and endogenous mutation rates in normal white blood cells. Led by Professor Sir Mike Stratton, Wellcome Sanger Institute
Patient and Public Engagement and Involvement
Alongside the scientific goals of the Mutographs project, we aim to add value to our research activity by engaging public and patient audiences with the research journey and its outputs and outcomes.
Our patient advocate representatives, Mimi and Maggie, along with science writer Kat Arney, will see behind the scenes of the Mutographs project and visit researchers, healthcare workers and cancer patients who are involved in the study at various locations around the world. They will explore peoples’ experiences of cancer and discover their motivations for being involved in research. They hope to use their findings to influence the direction of future research and improve the experience of patients involved in cancer studies. They also hope to highlight key research findings that may reveal opportunities to prevent cancer through changes in behaviour and policy in different regions.
Our first field trip is to Eldoret, Kenya, in February 2018. Eldoret is one of the collaborating medical centres where patient samples will be collected for the Mutographs project. It is in a region of Africa that has a high incidence of oesophageal squamous cell carcinoma. Our patient advocates will explore the patient experience of cancer and research in this region and feedback findings to the Grand Challenge team and wider public.
Contact: mutographsPPIE@sanger.ac.uk
Contact
If you need help or have any queries, please contact us using the details below.
External partners and funders
External
Paul Brennan, IARC - International Agency for Research on Cancer
Paul Brennan leads Work Package 1: Mutational signatures in five cancer types across five continents. This work supports Specific Aim 1: To elucidate the causes of major global geographical and temporal differences in cancer incidence through study of mutational signatures.
External
Ludmil Alexandrov, University of California, San Diego
Ludmil Alexandrov leads Work Package 2: Development of computational approaches for mutational signature analysis. This work supports Specific Aim 1: To elucidate the causes of major global geographical and temporal differences in cancer incidence through study of mutational signatures.
External
Allan Balmain, University of California, San Francisco
Allan Balmain leads Work Package 3: Mutational signatures of cancer development in rodents exposed to known or suspected carcinogens. This work supports Specific Aim 2: To identify and characterise the biological processes underlying mutational signatures.
External
David Phillips, King’s College London
David Phillips leads Work Package 4: Establishing a compendium of mutational signatures through exposure of cells in culture to defined mutagens. This work supports Specific Aim 2: To identify and characterise the biological processes underlying mutational signatures.
External
Peter Campbell, Wellcome Sanger Institute
Peter Campbell leads Work Package 5: Understanding mutation burdens and mutational signatures in non-cancer tissues. This work supports Specific Aim 3: To survey and monitor mutagenic exposures in normal cells in humans through mutational signature analysis.
External
Professor Sir Mike Stratton, Wellcome Sanger Institute
Mike Stratton leads Work Package 6: Surveying mutagenic exposures and endogenous mutation rates in normal white blood cells. This work supports Specific Aim 3: To survey and monitor mutagenic exposures in normal cells in humans through mutational signature analysis.
External
National Toxicology Program (NTP) Archives
A key data source will be rat and mouse tissue samples from the National Toxicology Program (NTP) Archives. The NTP Archives is a unique repository of rodent tumor tissues exposed to more than 590 chemical carcinogens. A thorough examination of these tissues will help scientists understand how various environmental exposures may play a role in cancer.
External
National Cancer Center (NCC), Japan
Contributor of sequenced data for colorectal, kidney, pancreatic, and oesophageal cancers, 700 sets in total, expanding the project’s coverage to East Asia. The NCC drives the nation’s research for personally optimized cancer treatments based on genomic information. With an edge in chemical carcinogenesis and cancer prevention, and the full spectrum of cancer research from prevention, diagnosis to treatment, the NCC brings insight to the project.
Related groups
Affiliated Sites
External
CRUK
External
COSMIC
External
King's College London
External
UC San Diego
External