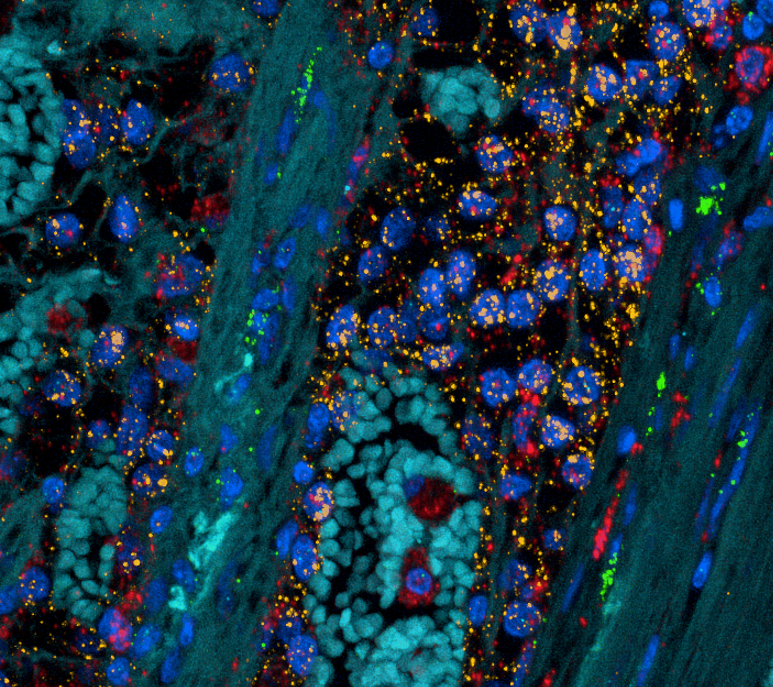
Microenvironment of kidney cancer
These studies are fundamental to better understand cancer biology as tumour behaviour is intricately dependent both on the oncogenic properties of cancer cells and their multi-cellular interactions.
Here we provide an overview of the data visualisation and links to download for over 270,000 single cell transcriptomes and spatial transcripomic data from this project.
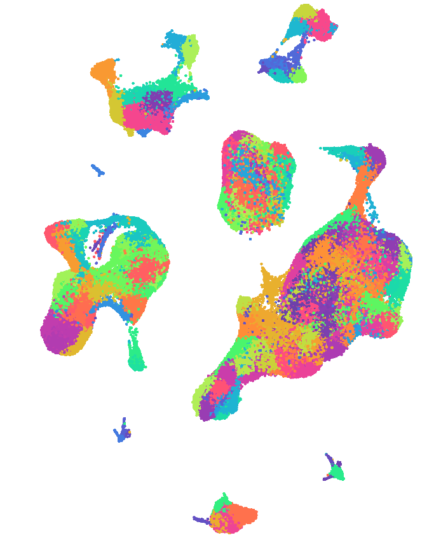
A comprehensive single cell atlas of kidney cancer
Over 270,000 single cell transcriptomes have been carefully annotated from 12 patients with kidney tumours. Cells were sampled from multiple regions within the tumour and the wider tumour microenvironment, including peripheral blood, the normal adjacent kidney, the tumour-normal interface, the adrenal gland, and any associated metastatic disease.
For the ~100,000 T cells captured, we have performed TCR sequencing, providing detailed insights into clonotypic evolution and expansion.
The data can be visualised via an interactive web-browser here.
Raw data can be downloaded from here.
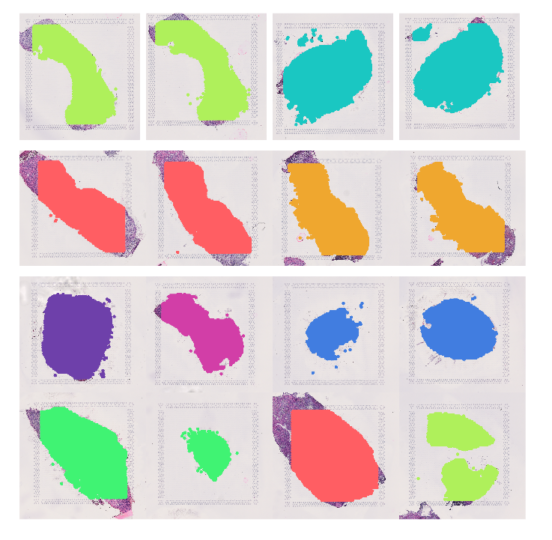
Spatial transcriptomic analysis at the tumour-normal interface
Kidney tumours are commonly enveloped with fibrous connective tissue that is the pseudocapsule or tumour-normal interface. It appears as though this layer constrains tumour growth and pseudocapsule invasion is correlated with tumour stage and grade. As part of this study, we compared the cellular make up of this interface with the tumour core.
Several notable differences are seen, including important interactions that are clinical targetable.
The data can be visualised via an interactive web-browser here.
Raw data can be downloaded from here.
External Contributors
External partners and funders
External
Cancer Research UK
External
