
Antarctic notothenioids fish genomes project
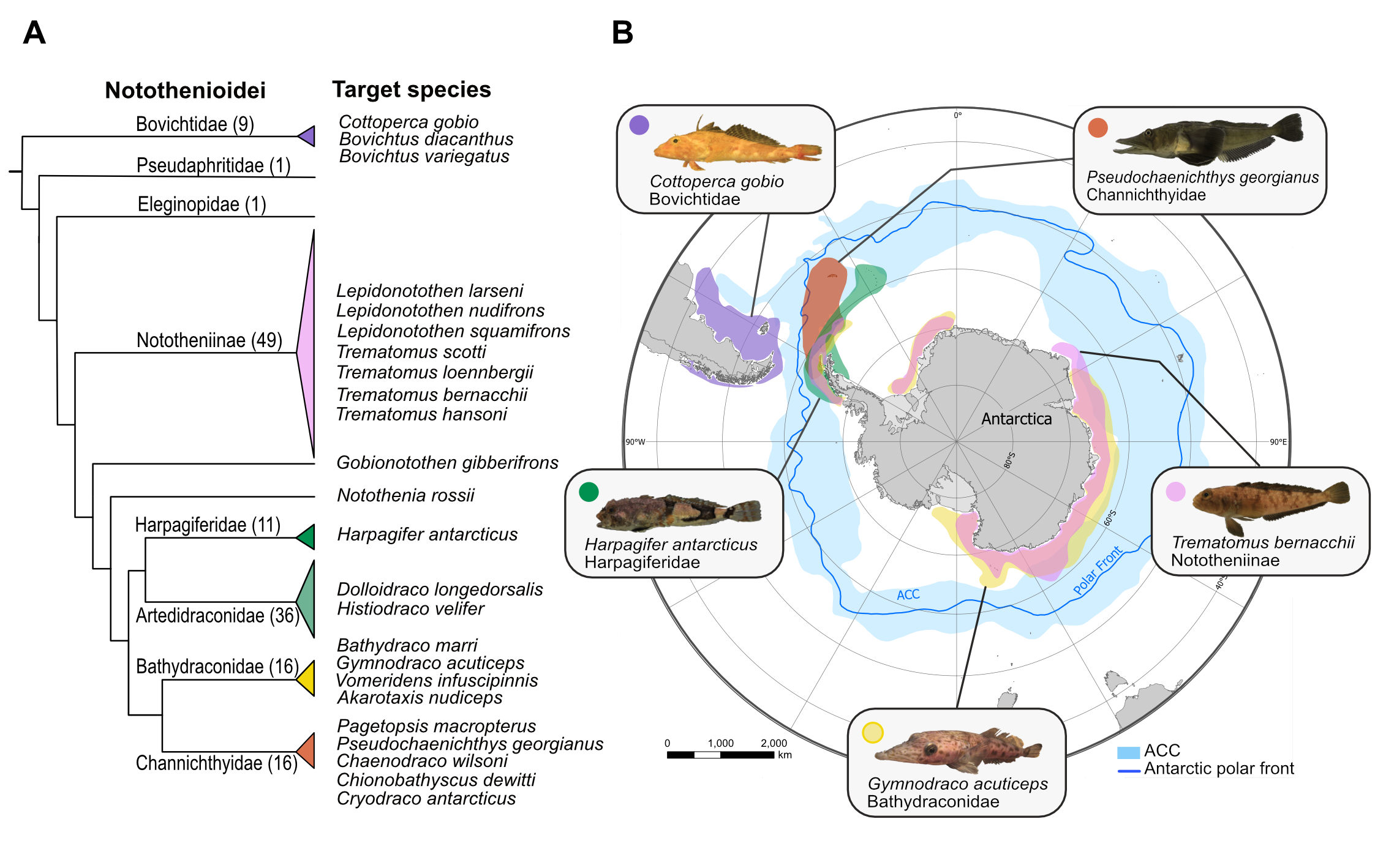
At the Wellcome Sanger Institute we are generating genome assemblies for 24 species across the notothenioid radiation. We are generating PacBio reference assemblies for five species, including the non-Antarctic Cottoperca gobio (synonym Cottoperca trigloides), and four cryonotothenioids; Trematomus bernachii, Harpagifer antarcticus, Gymnodraco acuticeps, and Pseudochaenichtys georgianus. Also, we are also generating draft reference assemblies for 20 more species.
This work is funded by the Wellcome Trust.
News
- Genomics of cold adaptations of the Antarctic notothenioid fish radiation. https://www.biorxiv.org/content/10.1101/2022.06.08.494096v1
- Cottoperca gobio reference assembly: The genome sequence of the channel bull blenny, Cottoperca gobio (Günther, 1861). https://wellcomeopenresearch.org/articles/5-148.
- Towards complete and error-free genome assemblies of all vertebrate species. https://www.nature.com/articles/s41586-021-03451-0
Data usage:
- These assemblies are provided by the Wellcome Sanger Institute and Cambridge University team of the Sanger Vertebrate Genomes Project
- The data under this project are made available subject to the Wellcome Sanger Institute Data Sharing policy (https://www.sanger.ac.uk/about/research-policies/open-access-science/)
Sanger people

Dr Richard Durbin
Associate Faculty
Previous Sanger people

Eric Miska
Associate Faculty/Group Leader
External partners and funders
External
William Detrich group
Northeastern University, College of Science
Professor Marine and Environmental Sciences, Marine Science Center